Scientists at the Center for Biomedical Imaging Innovation and Research have found that MRI signal can reveal microscopic features of axons, threadlike protrusions of nerve cells, that have until now been widely considered undetectable in vivo through non-invasive means. The finding, reported in the journal Communications Biology, has profound implications for research on and treatment of neurodegenerative conditions, such as Alzheimer’s disease or multiple sclerosis.
In the white matter of the the brain and the spinal cord, axons are packed in tight bundles that beam information among billions of neurons and link the nervous system with other major systems of the body. For the first time, scientists have demonstrated that variations in the caliber of the microscopic fibers that compose white matter can be detected with MRI. Such variations, also known as axonal beading, are a corollary of neuronal injury and have only been measurable in tissue samples removed from living organs, limiting research to preclinical methods such as post-mortem and animal studies. The new work combines theoretical advances, electron microscopy, novel computational modeling, and in vivo brain imaging of healthy controls and patients diagnosed with multiple sclerosis. The findings demonstrate that it is possible “to image cellular-level pathology with a clinically feasible MRI technique,” the authors write.
Although the resolution of magnetic resonance (MR) images is limited to the macroscopic scale of millimeters, the signal such images are reconstructed from originates at the level of molecules and has long tantalized scientists with the promise of delivering reliable information about structures far smaller than those visible on clinical MRI. Research into microstructure imaging (sometimes called microstructure mapping, microstructure modeling, or biophysical modeling of tissue microstructure) has embraced a technique called diffusion MRI, which emerged in the 1990s and enabled probing of cellular environments that heavily restrict natural random motion of water molecules. The method is most commonly used in brain imaging of white matter, where axons form tightly bundled tubular structures sheathed in a hydrophobic substance called myelin. Vividly color-coded tractographic maps that chart approximate orientations of axon bundles, for example, are computed from diffusion data.
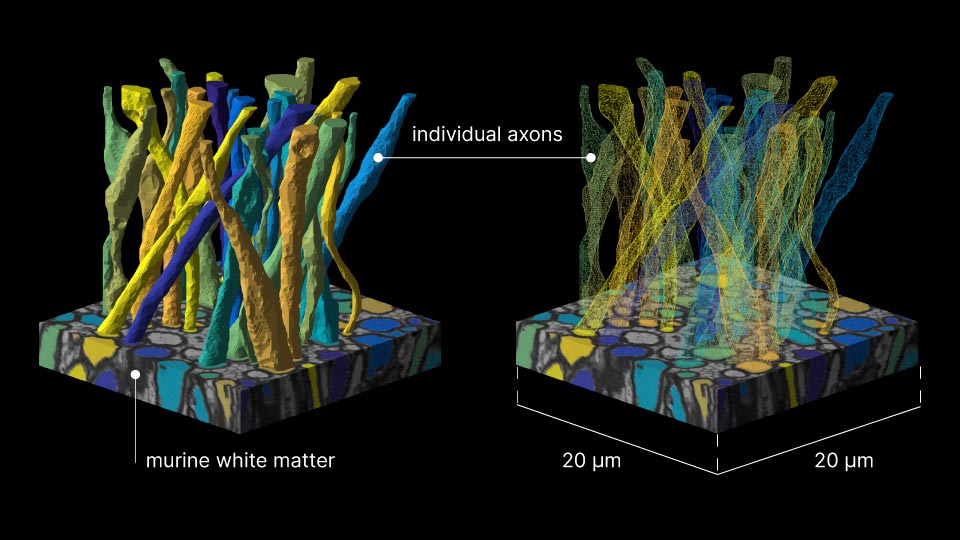
For more than a decade, microstructure models have idealized axons as “sticks,” infinitely narrow perfect cylinders.
“People always use this assumption without any doubt,” said Hong-Hsi Lee, MD, PhD, the lead author of the study. “But we doubt it very much because when we look at histology, it doesn’t look like that.”
The stick convention has endured because of fundamental lack of knowledge about what elements of axonal microenvironment give rise to which—if any—features of the diffusion MR signal. The basic unit of MRI measurement, a voxel, can be as small as one cubic millimeter—a sesame seed’s worth of tissue—but axons are thousands of times smaller, about one micrometer across.
Dmitry Novikov, PhD, senior coauthor of the study, compared detecting changes in axonal thickness inside an MRI voxel to finding a raised hand in a Manhattan city block. “How would you even be able to quantify exactly what happened?” he said. “If you look at histology of tissue, there are millions of parameters you could think of for describing its incredibly complex morphology.”
Plucking the signal features that reliably correspond to precise tissue-level phenomena from measurements coarser by three orders of magnitude has taken years of inquiry and ultimately called for a new way of investigating the microgeometry of white matter.
In 2014, Els Fieremans, PhD, senior coauthor of the study, and Dr. Novikov made theoretical predictions that brain diffusion MRI signal would exhibit a particular type of behavior incompatible with the stick model—a distinctive form of time-dependent decrease in the diffusion coefficient along axons. “It’s not something that you can just get out of the blue,” explained Dr. Novikov. “It’s a very specific functional form.”
In 2016, they observed such behavior in human brain studies but could not validate which features of tissue microstructure gave rise to it. To make the connection, they needed an experiment that would bridge tissue-level microgeometry with voxel-level diffusion MRI signal.
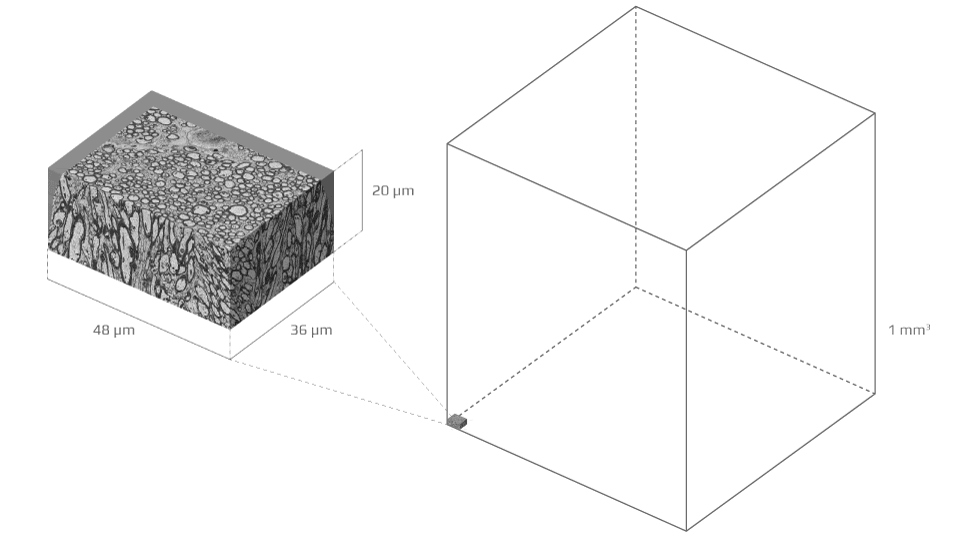
To build that bridge over three orders of magnitude in scales, the study team acquired a detailed electron microscopy sample of white matter—a tiny block of murine corpus callosum sliced into hundreds of sections, each a tenth of a micron thin—and segmented it in order to form meticulous three-dimensional digital replicas of 227 axons (the segmentation results were published in 2019).
Equipped with a ground-truth reference, the scientists went on to model diffusion MRI in silico—linking for the first time micro geometric features of axons to parameters of MRI measurements. Using the axon replicas, researchers created virtual axon test cases, each with selectively removed geometric features, such as mitochondria, diameter variations or undulation. Each of these cases was then subjected to diffusion MRI simulations. By systematically eliminating specific elements of the microscopic topology of axons and analyzing the resulting measurements, the researchers were able to discover which characteristic corresponds to changes in the very time-dependence they had predicted theoretically and observed in vivo years earlier. The relevant feature was variation in axonal diameter.
In order to establish the missing link between macro- and micro-scales, the team developed the first successful adaptation of Monte Carlo simulations to complex three-dimensional volumes obtained with electron microscopy—a unique technical achievement made possible by the kind of supercomputing power that is typically employed to solve the most challenging statistical problems or train artificial intelligence algorithms.
The researchers also took MRI brain scans of healthy volunteers and patients diagnosed with multiple sclerosis. In healthy volunteers, the scientists observed diffusion time-dependence patterns consistent with theoretically and numerically predicted effect of axon caliber variations. In multiple sclerosis patients, the study team found time-dependence parameters consistent with higher density of axonal caliber variation, indicative of pathological proliferation of mitochondria in MS lesions. Although this study focused on patients with MS, the findings may have implications also for other conditions, such as traumatic brain injury, associated through ex-vivo research with a phenomenon known as axonal beading and characterized by greater-than-usual variation in axonal diameters.
“The combination of theory, human experiment, and simulation is really unique,” said Dr. Fieremans, referring to the structure of the study. She noted that the findings mark a fundamental change in the understanding of microstructure imaging and its clinical applications. “We are revealing that there is an understudied axonal feature that you can access with diffusion MRI.”
Related Resources
Scripts for two-dimensional modeling of diffusion.
Data and code for precise, sample-based modeling of axon microgeometry.
Related Story
Hong Hsi Lee, alumnus of the Biomedical Imaging & Technology PhD Program at NYU Grossman School of Medicine, has just completed postdoctoral training at our Center and is headed to a postdoctoral fellowship at Massachusetts General Hospital. We take a look at his journey.



