A team of four machine learning researchers and 20 radiologists from the Center for Advanced Imaging Innovation and Research (CAI2R) and NYU Langone Health Department of Radiology has won an open competition to develop a deep learning network that detects lesions in digital breast tomosynthesis (DBT) data. The challenge, called DBTex for short, was organized by SPIE, the American Association of Physicists in Medicine (AAPM), the National Cancer Institute (NCI), and Duke AI in Radiology (DAIR). The three top entries were presented at SPIE Medical Imaging on February 16, 2021.
“Competitions are opportunities to test yourself,” said Krzysztof Geras, PhD, who led the group, dubbed NYU B-team, to first place. “When you train models for a publication, you can curate data and do a lot of things—consciously or unconsciously—to make your results look better,” said Dr. Geras. Challenges make such tweaks harder because entrants’ do not have access to test data. The winning team outranked the competition by combining innovative computer-science strategies, in-house data, and clinical expertise.
Why DBT?
Digital breast tomosynthesis is an imaging method that produces three-dimensional representations, or volumes, by combining images acquired from a range of angles.
“That’s important, because the breast is a three-dimensional object,” said radiologist Beatriu Reig, MD, MPH, member of the team. Dr. Reig explained that a volume provides much more information than does traditional two-dimensional digital mammography (DM), which creates “a single image where all those structures are superimposed on each other.”
The nuances matter. Linda Moy, MD, breast radiologists and member of the team, said that DBT “detects more cancers than a digital mammogram” and that it “reduces recall rates and false-positive rates,” which means fewer callbacks for additional imaging and biopsies.
However, the defining advantage of DBT—more and richer information—is also its costliest drawback. While DM produces four images (two views of each breast), DBT creates four volumes, each composed of 70 “slices” on average (volumes range from having just over a dozen to well over a hundred slices). About half of mammographies currently performed in the U.S. are done with DBT (all screenings at NYU Langone Health involve both DBT and DM).
“It takes a lot more time to read these studies,” said Dr. Moy. She estimates that the addition of DBT to the imaging protocol doubles the amount of time the fastest radiologists need to read an exam and more than triples the time for slower readers. “Your throughput really falls quickly.”
Artificial intelligence (AI) has the potential to reduce—perhaps eliminate—this bottleneck. “Wouldn’t it be great if we had some AI model that could tell me, ‘Hey, Linda, there’s something you should look at in slice number fourteen’,” Dr. Moy said. She added that the possible benefits of AI are far greater for DBT than for DM, but there have only been a handful of peer-reviewed studies exploring deep learning in DBT.
DBT Data: Too Large and Not Enough
The relative dearth of machine learning research on DBT stems from two difficulties: the scarcity of high-quality training data—an ironic consequence of the amount of information created by this modality; and DBT’s sheer size, which calls for huge amounts of computational power.
Annotation of reams of DBT volumes is expensive as it diverts radiologists’ attention from patient cases that generate clinical revenue. “It’s a very time intensive process and it requires someone who’s very well trained in breast imaging,” said Dr. Reig. Because screening aims at catching subclinical disease long before the development of palpable masses, “the findings are often pretty subtle,” she said.
The DBTex competition dataset itself exemplified this difficulty. The data comprised about 1,000 exams (3,500 volumes). Meanwhile, outside of medicine, gold-standard datasets of natural images—pictures of animals, people, and objects—contain many millions of distinct examples.

Researchers on NYU Langone’s team supplemented the challenge dataset with their own, larger caches. A contingent of radiologists from the breast imaging division—led by Dr. Reig, Dr. Moy, and Laura Heacock, MD—annotated biopsy-proven DBT studies from a set of 100,000 exams (400,000 volumes). For each volume, readers reviewed the findings, precisely highlighted lesions that led to biopsies, consulted results of those biopsies, and labeled the lesions accordingly as benign or malignant. In addition, the scientists tapped an in-house set of about 200,000 DM exams (one million DM images) they had built for prior research.
“It’s one of the crucial advantages that we have,” said Jungkyu Park, MS, machine learning researcher on the team and doctoral candidate in the Biomedical Imaging & Technology PhD Training Program at NYU Grossman School of Medicine, referring to the data. “Without them, we wouldn’t have been able to win,” he said, but the homegrown stock of annotated images was not the be-all and end-all of the team’s success.
“Even after adding NYU data, there’s still not a lot of images that have lesions,” Park said. To squeeze more learning out of their image sets, the researchers applied subtle transformations—such as scaling, rotation, or shearing. Doing so provided more variations of lesion-positive images to the networks.
The researchers also increased the variety of learning with a technique called model ensembling. They created 40 separate machine learning sub-models, each of which underwent training and validation on slightly different subsets of data. Then, they tested each sub-model and “ensembled” the resulting predictions. “It’s a very well known technique from training classification models,” said Dr. Geras. The various sub-models make different outlier predictions, many of which are erroneous. Ensembling removes those outliers and amplifies the predictions that most of the sub-models agree on. “This leads to a much stronger model,” Dr. Geras said.
“Another characteristic of DBT volume is that image sizes are very large,” said Park. A DBT slice is between twice and eighteen times as large as the natural images used to train some of the state-of-the-art deep neural networks. The stark size differences stem directly from the nature of high-resolution DBT, which radiologists comb through in search of small suspect areas. Moreover, compressing DBT threatens the loss of medically relevant information. While a thumbnail picture of a bus may be clear enough for the vehicle to remain recognizable, a heavily downsampled DBT slice risks no longer showing the diaphanous whisp of problematic tissue.
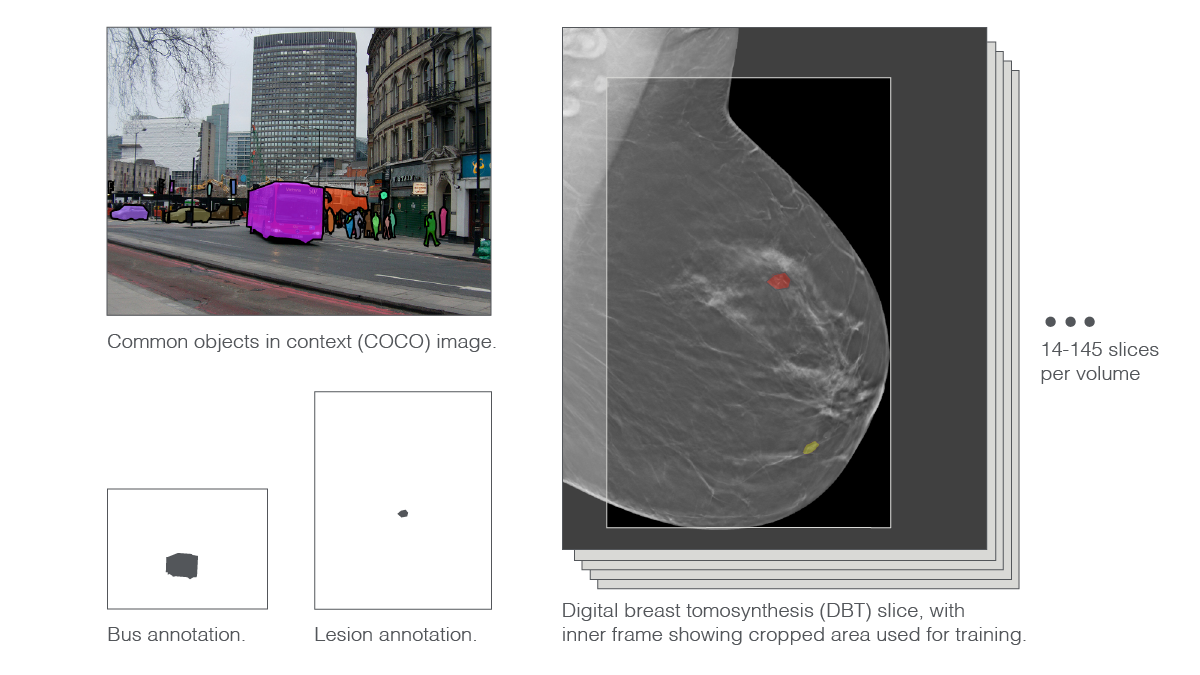
To scale things down a bit, researchers cropped away background areas and trimmed the very top and bottom parts of the images. Then, they slightly downsampled the resulting pictures. Even with these preprocessing maneuvers, the scientists had to push their supercomputer’s 144 GPUs to the limit.
“We used such tricks as mixed precision training,” said Dr. Geras, referring to a method that allows fitting more data and a bigger neural net on a GPU by storing some numbers in 16-bit representations and saving the standard 32-bit representations for the numerical operations that require more precision. Researchers also trained on multiple GPUs simultaneously and used so-called large model support, a property of high-performance computing clusters in which GPUs have access to the computer’s main memory. Such configuration enables computational loads that exceed the capacity of the GPU by temporarily drawing on the main memory. “You have to understand your hardware really well,” said Dr. Geras.
The AI Sees It Differently
DBT data can sometimes seem ambiguous to an AI—such as when one lesion appears behind another on the same slice. DBT does not take a 360-degree view of the breast, hence in some cases there are not enough projections to precisely resolve an apparent overlap.
“It’s a little bit different for humans,” said Dr. Reig, explaining that this tends to be less of a problem for radiologists. “All lesions are three dimensional objects and there’s never one that’s on a single slice,” she said. But for the deep learning model, the ambiguity caused a drag on performance. Park said that the phenomenon “makes it difficult to determine the true spacial extent of the lesion.”
To straighten out this wrinkle, researchers created an algorithm that defines hundreds of rectangular “anchors” in each slice of a DBT volume. The anchors are identical across all slices, but the model makes distinct predictions associated with each anchor on each slice. The model also assigns a confidence level to each prediction. This allows the software to focus on any particular anchor and look through all predictions associated with it throughout the entire volume. The most confident predictions are retained, the rest discarded. “Doing this, we figure out which slice the lesion is centered in,” said Park, adding that the strategy made a marked improvement in the team’s score. “With finding the center slice, the mean sensitivity of finding the lesion at one, two, three, and four false-positives per volume”—a key metric in the challenge—“increased by about 0.1.” With additional tweaks, the model ultimately scored above 0.9.
‘B’ on a Team
Challenges like DBTex require technical expertise, computing infrastructure, and data—but also teamwork. NYU B-team brought together research faculty, graduate students, postdoctoral fellows, radiology residents, radiology fellows, and clinical faculty. At one point during the SPIE Medical Imaging challenge session, where top three entrants presented their work, the moderator and challenge co-organizer Maciej Mazurowski, PhD, said, “If I am reading the name correctly, then I wonder what the A-team is.”
The comment prompted speculation that the ‘B’ stands for ‘Breast,’ but Dr. Geras said that “the truth is nobody really knows what the ‘B’ in ‘B-team’ is.” Regardless, team members pointed to the collaboration between machine learning scientists and clinical radiologists as the kernel of their success.
“The benefit is mutual,” said Dr. Geras. “Working together is valuable both for us—helping us understand these data and obtain them in greater amounts—and for them, because it helps them better understand what we do and better prepare for what will be the future of radiology.”
Dr. Reig echoed this sentiment. “We want to improve on a radiologist’s eyes and experience,” she said. “The idea is that down the line this is going to help doctors and patients.”
Related Stories
Yiqiu "Artie" Shen, machine learning researcher who develops artificial intelligence systems for medical imaging, talks about AI's ability to explain itself, guide discovery, and predict cancer risk.
Krzysztof Geras and Jan Witowski, machine learning researchers in medical imaging, talk about understanding one's data, working across disciplines, and radiologists' "new colleague."
A combination of radiologists and AI reduced overdiagnosis and more accurately identified breast cancer in ultrasound exams.



